Tipping-Point Detection
Bulk data analysis
Detecting tipping point (deterioration) based on bulk data
Please input data on the card below
The uploaded dataset contains 0 columns. Please indicate the information of samples and time points.
There are 1 time points/ stages.
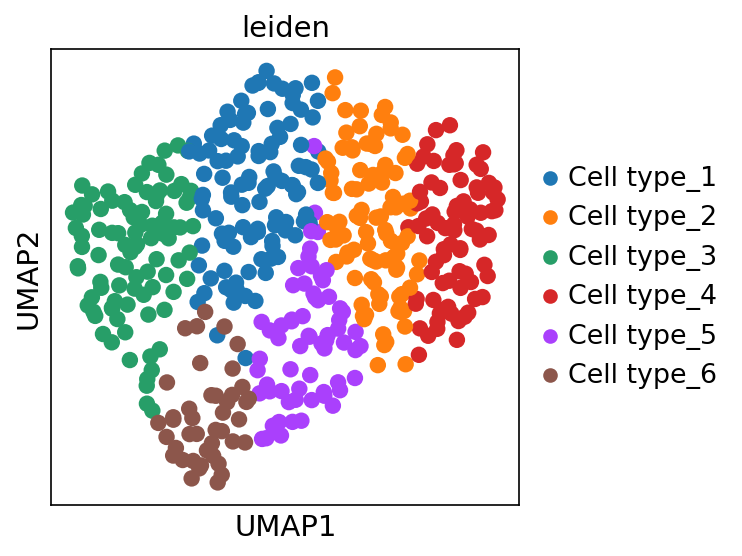
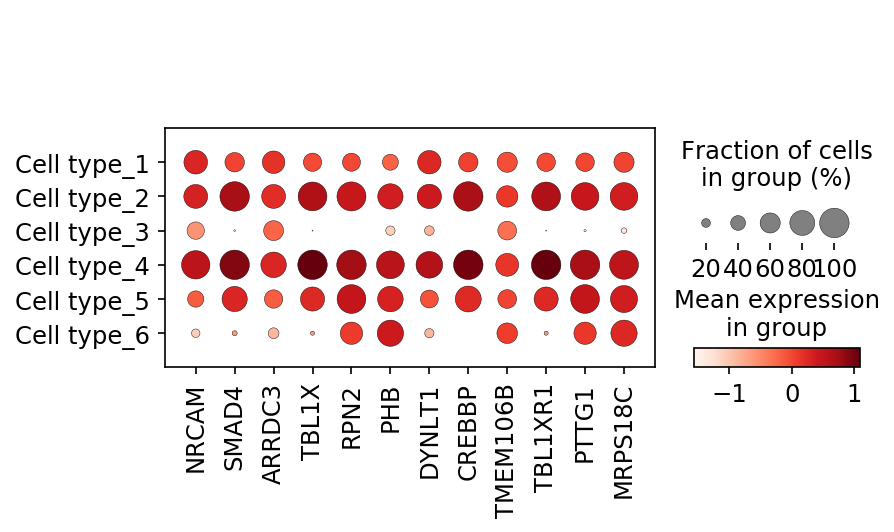
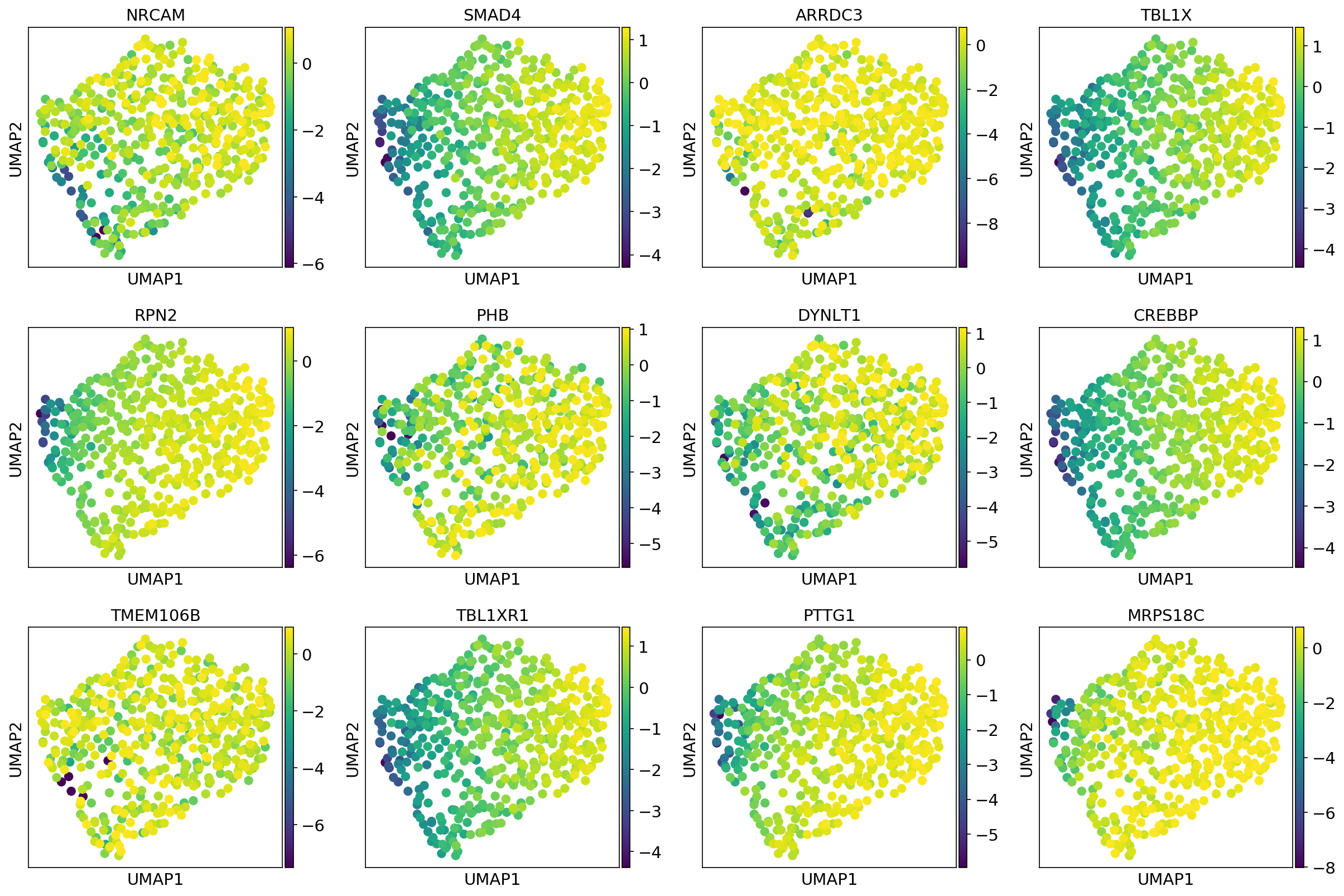
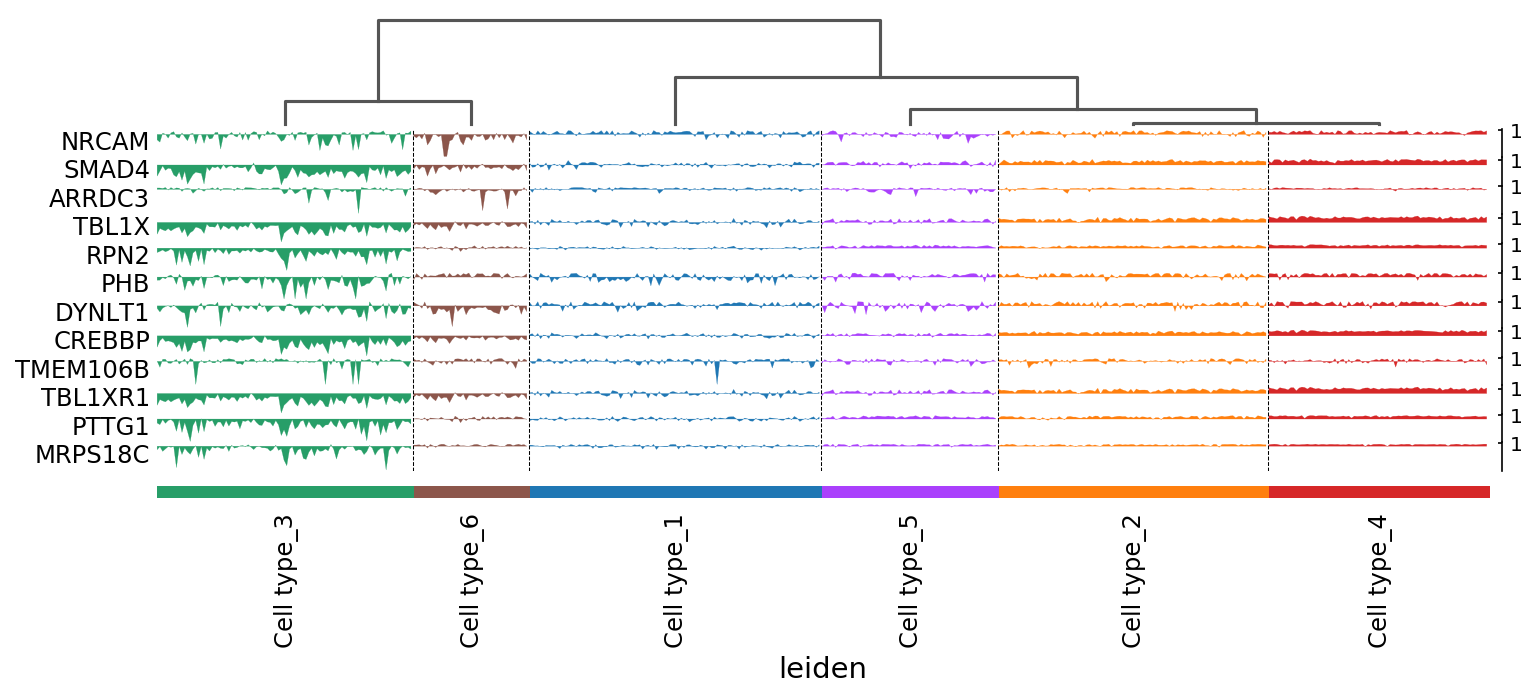
Single-cell data analysis
Detecting tipping point (cell fate commitment) based on single-cell data
Please input data on the card below
The uploaded dataset contains 0 columns. Please indicate the information of samples and time points.
There are 1 time points/ stages.