Ⅰ. What is Thyroid carcinoma (TCGA_COAD & Homo sapiens)
or
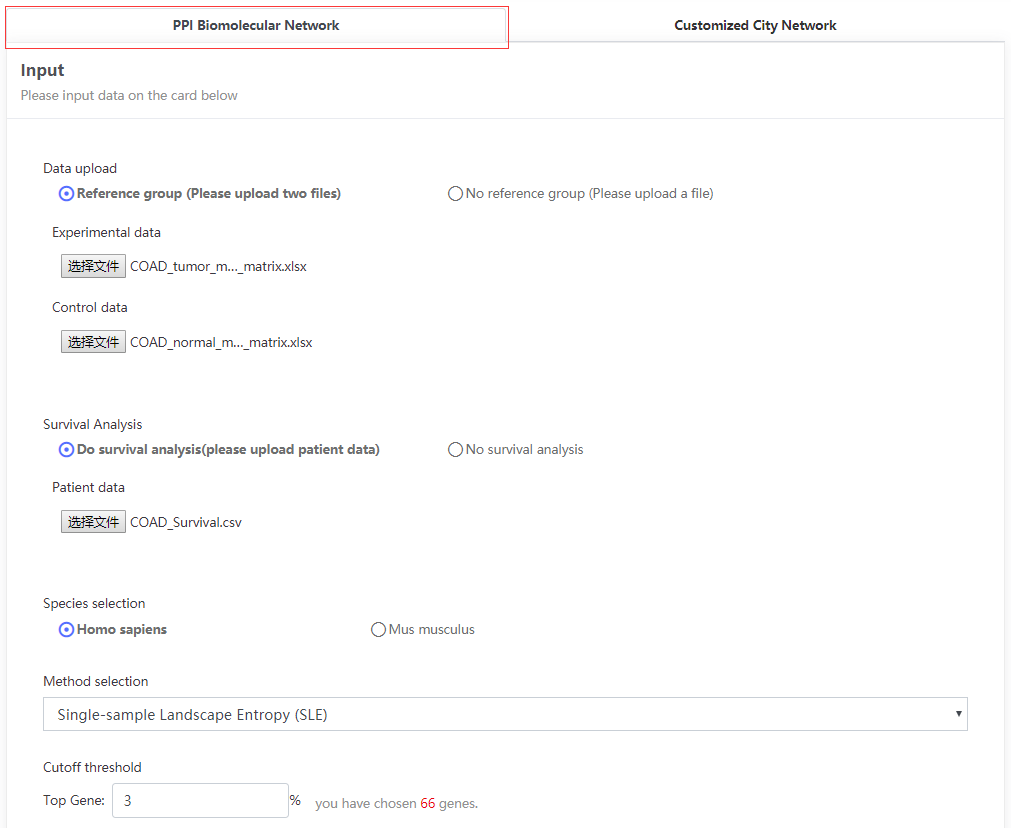
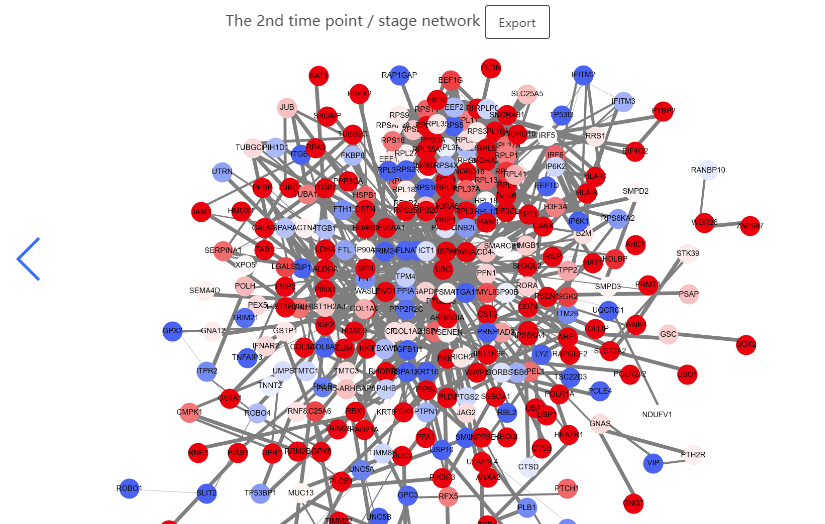
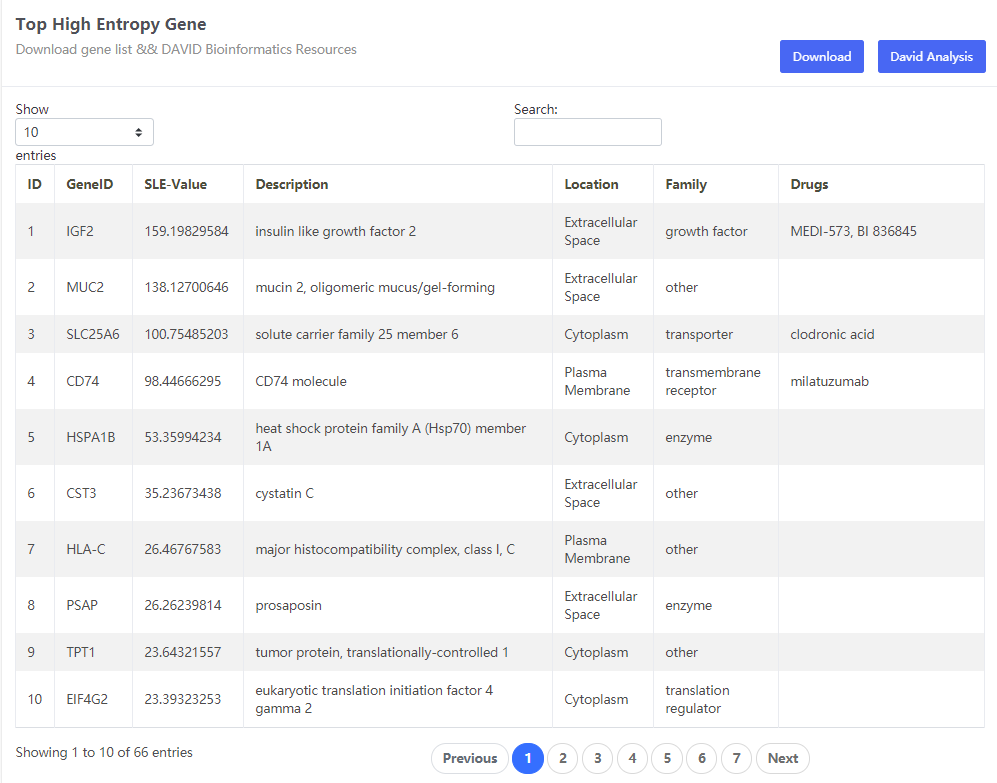
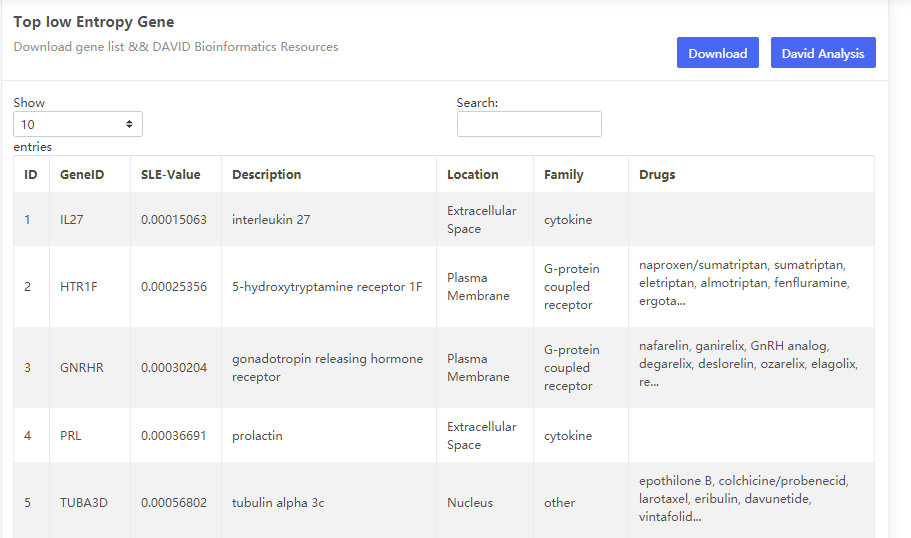
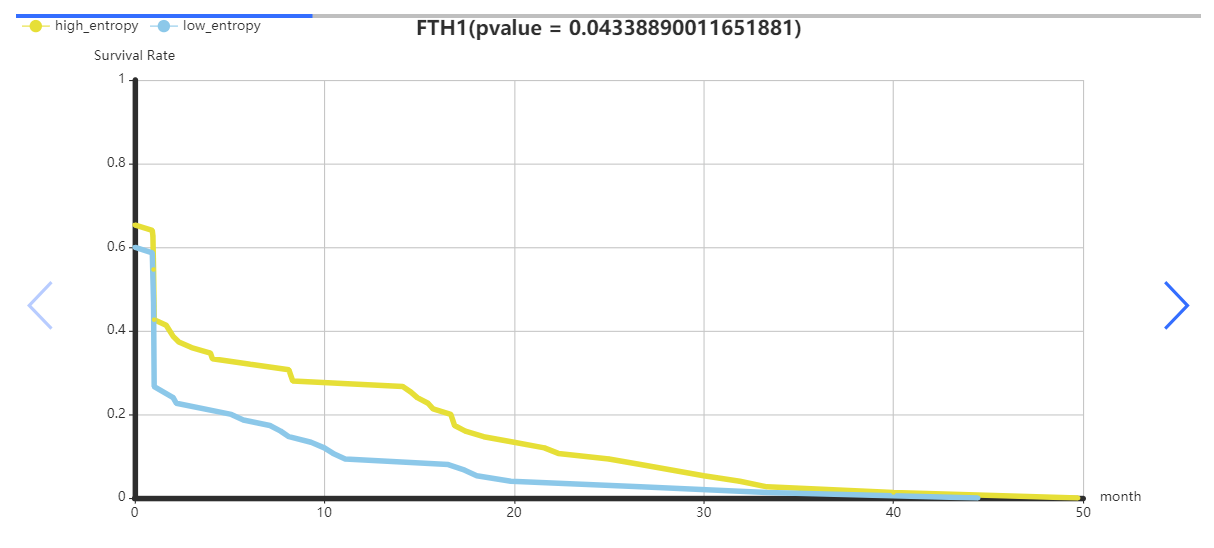
This data set is applicable to PPI Biomolecular Network.
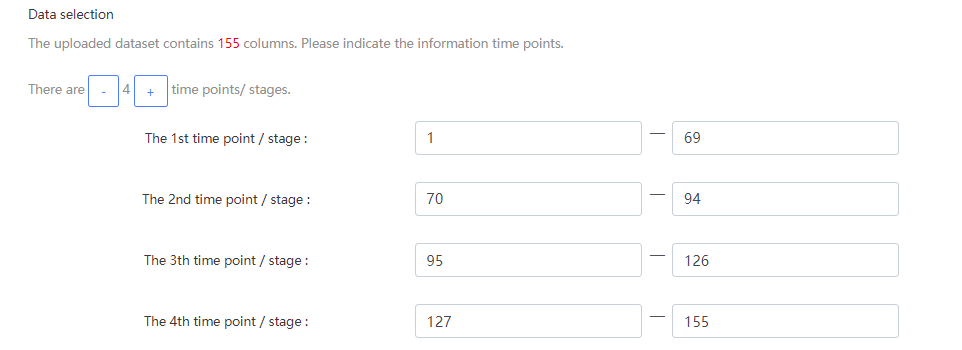
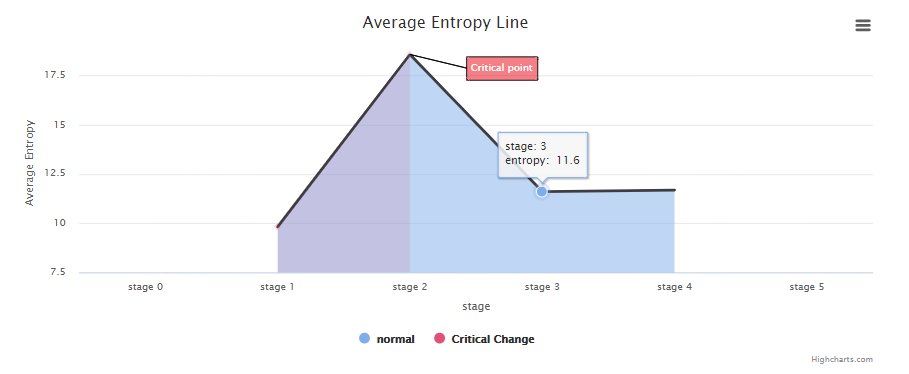
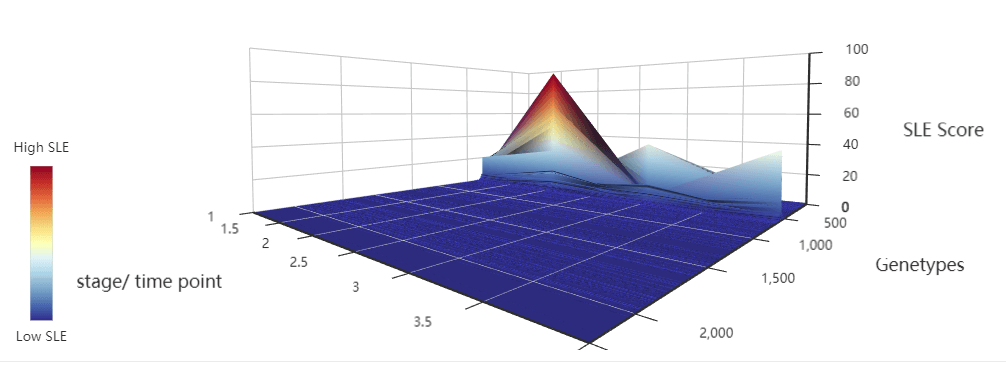
The cancers datasets from TCGA contained RNA-Seq data and included both tumor and tumor-adjacent samples. The tumor samples were divided into different stages according to corresponding clinical information of TCGA. (https://portal.gdc.cancer.gov/projects/TCGA-COAD)